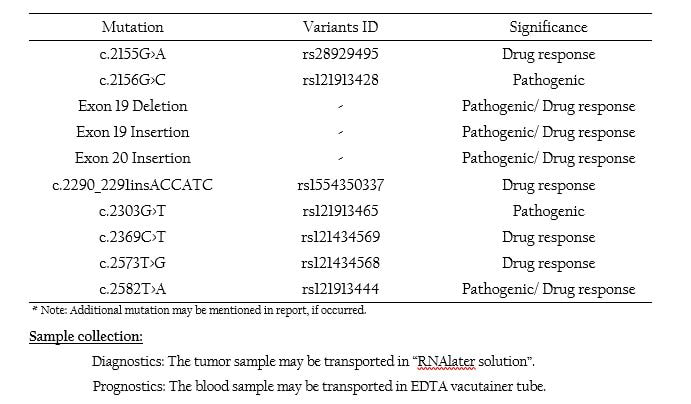
Lung Cancer- EGFR Mutation Panel
EGFR overview:
- Epidermal growth factor receptor (EGFR) belongs to a family of receptor tyrosine kinases (RTKs) that include EGFR/ERBB1, HER2/ERBB2/NEU, HER3/ERBB3, and HER4/ERBB4.
- The binding of ligands, such as epidermal growth factor (EGF), induces a conformational change that facilitates receptor homo- or heterodimer formation, thereby resulting in activation of EGFR tyrosine kinase activity.
- Activated EGFR then phosphorylates its substrates, resulting in activation of multiple downstream pathways within the cell,
- Including the PI3K-AKT-mTOR pathway involved in cell survival, and the RAS-RAF-MEK-ERK pathway involved in cell proliferation.
- Lung Cancer is one of the major cancer deaths and there is an estimated incidence of 1.2 million new cases each year worldwide[1]
- Deletion is the most common mutations in EGFR and has been observed in a number of lung carcinoma[2]
- The effects of smoking on the survival rate of the lung cancer patients proved to be a poor prognostic factor[3]
- It has been confirmed that non-smokers were associated with a significantly higher EGFR mutation rate[4]
- EGFR mutation is the most common oncogenic driver in Asian Non-small cell lung carcinoma (NSCLC) patients and women are more susceptible for EGFR mutations[5]
- The EGFR-tyrosine kinase inhibitors (TKIs), gefitinib and erlotinib offer better efficacy and quality of life for lung cancer patients[6] and have hence emerged as an important frontline therapy for patients with EGFR-mutant, non-small cell lung cancer[7].
- Some studies have shown that EGFR expression in NSCLC is associated with reduced survival[8], frequent lymph node metastasis and poor chemosensitivity[9].
- EGFR is expressed in up to 93% of NSCLC patients in whom approximately 45% show overexpression[9].
- Approximately 10% of patients with NSCLC in the US and 35% in East Asia have tumor associated EGFR mutations[10&11].
- The mutations occur within EGFR exons 18-21, which encodes a portion of the EGFR kinase domain.
- Approximately 90% of these mutations are exon 19 deletions or exon 21 point mutations[12] These mutations increase the kinase activity of EGFR, leading to hyperactivation of downstream pro-survival signaling pathways[13].
- About 15–30% of NSCLCs harbour activating mutations in codons 12 and 13 of the KRAS gene[14]
- Mutations in KRAS have been proposed as a mechanism of primary resistance to gefitinib and erlotinib
- The EGFR has been found in the pathogenesis of multiple human tumours and higher levels of expressions of this receptor have been found in 30–40% of breast carcinomas and appears to confer a worse prognosis[15].
- Overexpression of the receptor has also been noted frequently in breast, bladder, and ovarian tumours, and in various squamous carcinomas[15].
- The EGFRvIII stimulates cell proliferation independently of ligand interaction[16] and enhances the tumourigenicity[17].
- GLOBOCAN. Cancer incidence, mortality and prevalence worldwide (2000 estimates); 2001.
- Garcia de Pallazzo I, et al. (1993). Cancer Res, 53: 3217-20.
- Ferketich AK, et al. (2013). Cancer, 119: 847-53
- Ren JH, et al. (2012). Environ Mol Mutagen, 53:78-82
- Hsu KH, et al. (2015) PLoS One, 10:e0120852.
- Greenhalgh J, et al. (2016). Cochrane Database Syst Rev, CD010383
- Reck M, et al. (2013). Lancet, 382:709–19
- Scagliotti GV, et al. (2004). Clin Cancer Res, 10:4227–32
- Fontanini G, et al. (1998). Clin Cancer Res, 4:241-9.
- Lynch TJ, et al. (2004). N Engl J Med, 350(21):2129-39.
- Paez JG, et al. (2004). Science, 304(5676):1497-500.
- Ladanyi M1, and Pao W. (2008). Mod Pathol. 21(2):S16-22.
- Sordella R1, et al. (2004). Science. 305(5687):1163-7.
- Mitsudomi, T. et al. (1991). Cancer Res. 51, 4999-5002.
- Harris AL., et al. (1992). Natl. Cancer Inst. Monogr, 11:181-187.
- Ekstrand AJ, et al. (1994). Oncogene, 9: 2313-20.
- Nishikawa R, et al. (1994). Proc Natl Acad Sci USA, 91: 7727-3128.