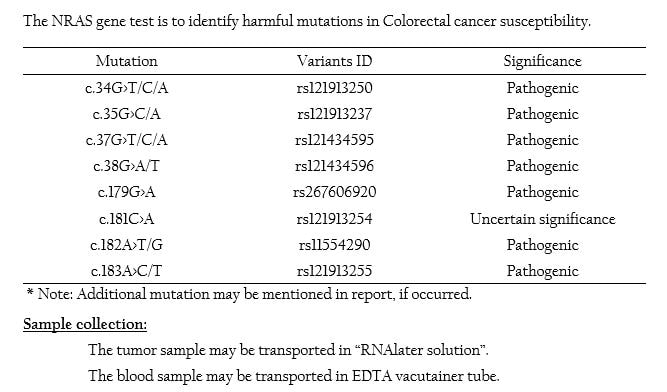
NRAS Mutation Panel
NRAS overview:
- The RAS genes are highly homologous but functionally distinct[1]
- RAS proteins are central mediators that downstream the growth factor receptor signaling and are critical for cell proliferation, survival, and differentiation.
- RAS can activate several downstream effectors, including the PI3K-AKT-mTOR pathway, which is involved in cell survival, and the RAS-RAF-MEK-ERK pathway, which is involved in cell proliferation.
- Activating mutations within the RAS gene result in constitutive activation of the RAS GTPase, even in the absence of growth factor signaling which results in sustained proliferation signal within the cell.
- Specific RAS genes are recurrently mutated in different malignancies. RAS mutations are particularly common in colon cancer, lung cancer, and pancreatic cancer[2].
- NRAS are closely related RAS oncogene family members, and mutations in genes either at codons 12, 13 (exon 2), codon 61 (exon 3) and codon 146 (exon 4) result in overactive RAS signalling which promotes oncogenesis.
- In majority of cases, NRAS mutations are missense mutations that introduce an amino acid substitution at position 61.
- The result of these mutations is constitutive activation of NRAS signalling pathways.
- NRAS mutations seem to arise at a later stage in the development of malignancy, unlike KRAS mutations, which arise early[3].
- NRAS mutations are now regularly identified in the clinical treatment of mCRC as part of routine RAS testing prior to EGFR inhibitor therapy.
- About 2-7% of the NRAS mutations are known to harbour in metastatic colorectal cancer (mCRC) and the frequency of NRAS mutations in colorectal cancer (CRC) is about 2% to 4%[4&5].
- KRAS mutations in colorectal cancers are seen in right-sided tumours, while NRAS mutations are seen in left-sided tumours and mostly in females[4].
- Patients with CRC who do not have mutations in both KRAS and NRAS appear to benefit from anti-EGFR mAb therapy[6&7].
- Patients with CRC are recommended for genetic testing of KRAS or NRAS mutations due to fact that, presence of KRAS or NRAS mutations may fail the effects of anti-EGFR mAb therapies [8&9].
- There were no significant differences found in terms of prognosis of mutated NRAS and wild-type NRAS[10&11].
- Recent studies have proven that, mutated NRAS protects the CRC cells from stress induced apoptosis, as a result, mutated NRAS provides a steady-state survival signal and contributed to cancer development, progression and also promotes inflammation[12].
- Unlike KRAS mutations, NRAS mutations seem to arise at a much later stage in the development of malignancies[13].
- The frequency of NRAS mutations and their relationship to clinical, pathologic, and molecular features remains uncertain till date[4].
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. (2011). Nat Rev Cancer, 13:(11):761-74.
- Karnoub AE and Weinberg RA. (2008). Nat Rev Mol Cell Biol, 9(7):517-31.
- Demunter A, et al., (2001). J Invest Dermatol, 117:1483-1489.
- Irahara N, et al., (2010). The American journal of surgical pathology, part B, 19:157-63.
- Douillard JY, et al., (2013). N Engl J Med. 369:1023-34.
- Ciombor KK, et al., (2015). Annu Rev Med, 66: 83-95.
- Amado RG, et al., (2008). J Clin Oncol, 26:1626-1634.
- Allegra CJ, et al., (2009). J Clin Oncol, 27: 2091-6.
- De Roock W, et al., (2010). Lancet Oncol, 11: 753-62.
- Mouradov D, et al., (2013). Am J Gastroenterol, 108: 1785-93.
- Gavin PG, et al., (2012). Clin Cancer Res. 18: 6531-41.
- Wang Y, et al., (2013). Cancer Discov, 3: 294- 307.
- Demunter A, et al., (2001). J Invest Dermatol, 117:1483-1489.